Analyzing network scale-up data using the networkreporting package
Dennis M. Feehan
2026-02-25
network_scaleup.RmdIntroduction
The networkreporting package has several tools for analyzing survey data that have been collected using the network scale-up method.
This introduction will assume that you already have the networkreporting package installed. If you don’t, please refer to the introductory vignette (“getting started”) for instructions on how to do this.
Review of the network scale-up method
For the purposes of this vignette, we’ll assume that you have conducted a survey using network scale-up questions in order to estimate the size of a hidden population. Analytically, using the scale-up estimator involves two steps:
- step 1: estimating the size of the survey respondents’ personal networks (their degrees)
- step 2: estimating the size of the hidden population by combining the estimated network sizes (from step 1) with the number of connections to the hidden population
We’ll quickly review each of these steps, and then we’ll show how to use the package to carry the estimation out.
Step 1: estimating network sizes
Here, we will use the known population estimator for respondents’ degrees (Killworth et al., 1998; Feehan and Salganik, 2016). In order to estimate the degree of the th survey respondent, we use
where is the total size of the population, is the size of the th population of known size, and is the number of connections that survey respondent reports between herself and members of the th population of known size.
Step 2: estimating hidden population sizes
Once we have the estimates of the respondents’ degrees, we use them to produce an estimate for the size of the hidden population:
where is the size of the population of interest (which we want to estimate), is the set of respondents in our sample, and is the estimate of the size of respondent ’s degree, obtained using the known population method.
Preparing data
In order to use the package, we will assume that you start with two datasets: the first is a survey containing information collected from respondents about their personal networks; the second is information about the sizes of several populations.
The example data for this vignette are provided with the
networkreporting package, and can be loaded by typing
library(networkreporting)
library(surveybootstrap)
## column names for connections to hidden population numbers
hidden.q <- c("sex.workers", "msm", "idu", "clients")
## column names for connections to groups of known size
hm.q <- c("widower", "nurse.or.doctor", "male.community.health", "teacher",
"woman.smoke", "priest", "civil.servant", "woman.gave.birth",
"muslim", "incarcerated", "judge", "man.divorced", "treatedfortb",
"nsengimana", "murekatete", "twahirwa", "mukandekezi", "nsabimana",
"mukamana", "ndayambaje", "nyiraneza", "bizimana", "nyirahabimana",
"ndagijimana", "mukandayisenga", "died")
## size of the entire population
tot.pop.size <- 10718378The example data include two datasets: one has all of the responses from a network scale-up survey, and the other has the known population sizes for use with the known population estimator.
Preparing the known population data
The demo known population data are in
example.knownpop.dat:
example.knownpop.dat## known.popn size
## 1 widower 36147
## 2 nurse.or.doctor 7807
## 3 male.community.health 22000
## 4 teacher 47745
## 5 woman.smoke 119438
## 6 priest 1004
## 7 woman.gave.birth 256164
## 8 muslim 195449
## 9 incarcerated 68000
## 10 man.divorced 50698
## 11 nsengimana 32528
## 12 murekatete 30531
## 13 twahirwa 10420
## 14 mukandekezi 10520
## 15 nsabimana 48560
## 16 mukamana 51449
## 17 ndayambaje 22724
## 18 nyiraneza 21705
## 19 bizimana 38497
## 20 nyirahabimana 42727
## 21 ndagijimana 37375
## 22 mukandayisenga 35055example.knownpop.dat is very simple: one column has a
name for each known population, and the other has its toal size. We
expect that users will typically start with a small dataset like this
one. When using the networkreporting package, it is more
useful to have a vector whose entries are known population sizes and
whose names are the known population names. The df.to.kpvec
function makes it easy for us to create it:
kp.vec <- df.to.kpvec(example.knownpop.dat, kp.var="known.popn", kp.value="size")
kp.vec## widower nurse.or.doctor male.community.health
## 36147 7807 22000
## teacher woman.smoke priest
## 47745 119438 1004
## woman.gave.birth muslim incarcerated
## 256164 195449 68000
## man.divorced nsengimana murekatete
## 50698 32528 30531
## twahirwa mukandekezi nsabimana
## 10420 10520 48560
## mukamana ndayambaje nyiraneza
## 51449 22724 21705
## bizimana nyirahabimana ndagijimana
## 38497 42727 37375
## mukandayisenga
## 35055Finally, we also need to know the total size of the population we are making estimates about. In this case, let’s assume that we’re working in a country of 10 million people:
# total size of the population
tot.pop.size <- 10e6Preparing the survey data
Now let’s take a look at the demo survey dataset, which is called
example.survey:
head(example.survey)## id cluster region indweight sex age.cat widower nurse.or.doctor
## 1 1.1.1 1 Kigali City 0.330602 Male [25,35) 3 2
## 2 1.1.2 1 Kigali City 0.330602 Male [25,35) 0 2
## 3 1.1.3 1 Kigali City 0.330602 Male [25,35) 2 8
## 4 1.1.4 1 Kigali City 0.330602 Male [25,35) 0 1
## 5 1.1.5 1 Kigali City 0.330602 Male [25,35) 0 0
## 6 1.1.6 1 Kigali City 0.330602 Male [25,35) 7 4
## male.community.health teacher woman.smoke priest civil.servant
## 1 1 5 1 0 5
## 2 1 5 0 0 5
## 3 0 3 0 0 50
## 4 0 0 0 0 5
## 5 0 0 0 0 5
## 6 0 8 2 0 6
## woman.gave.birth muslim incarcerated judge man.divorced treatedfortb
## 1 3 4 2 3 2 0
## 2 3 0 2 3 1 0
## 3 4 3 0 0 2 0
## 4 0 0 0 0 0 0
## 5 0 0 0 0 1 0
## 6 3 4 3 0 1 0
## nsengimana murekatete twahirwa mukandekezi nsabimana mukamana ndayambaje
## 1 0 0 2 1 2 3 1
## 2 3 2 0 0 1 0 0
## 3 0 0 0 1 2 0 0
## 4 1 0 0 0 0 0 0
## 5 0 0 0 0 0 0 0
## 6 1 1 0 0 0 0 0
## nyiraneza bizimana nyirahabimana ndagijimana mukandayisenga died sex.workers
## 1 0 2 0 1 0 0 0
## 2 0 0 0 0 0 1 0
## 3 0 0 0 0 0 2 0
## 4 0 0 0 0 0 0 0
## 5 0 0 0 0 0 0 0
## 6 0 0 0 0 0 4 0
## msm idu clients
## 1 0 0 2
## 2 0 0 1
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 10The columns fall into a few categories:
- an id variable for each respondent:
id - information related to the sampling design of the survey:
cluster,region, andindweight. - demographic characteristics of the respondents:
sexandage.cat - responses to questiona bout populations whose total size is known:
widower, …,mukandayisenga - questions about hidden populations:
died, …,clients
This is the general form that your survey dataset should have.
Topcoding
Many network scale-up studies have topcoded the responses to the aggregate relational data questions. This means that researchers considered any responses above a certain value, called the topcode, to be implausible. Before proceeding with the analysis, researchers substitute the maximum plausible value in for the implausible ones. For example, in many studies, researchers replaced responses with the value 31 or higher with the value 30 before conducting their analysis (see Zheng, Salganik, and Gelman 2006).
We won’t discuss whether or not this is advisable here, but this is
currently a common practice in scale-up studies. If you wish to follow
it, you can use the topcode.data function. For example,
let’s topcode the responses to the questions about populations of known
size to the value 30. First, we’ll examine the distribution of the
responses before topcoding:
## make a vector with the list of known population names from
## our dataset of known population totals
known.popn.vars <- paste(example.knownpop.dat$known.popn)
## before topcoding: max. response for several popns is > 30
summary(example.survey[,known.popn.vars])## widower nurse.or.doctor male.community.health teacher
## Min. : 0.0000 Min. : 0.0000 Min. : 0.000 Min. : 0.000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.000 1st Qu.: 0.000
## Median : 0.0000 Median : 0.0000 Median : 0.000 Median : 0.000
## Mean : 0.6101 Mean : 0.5112 Mean : 0.724 Mean : 1.356
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000 3rd Qu.: 1.000 3rd Qu.: 1.000
## Max. :95.0000 Max. :40.0000 Max. :95.000 Max. :95.000
##
## woman.smoke priest woman.gave.birth muslim
## Min. : 0.000 Min. : 0.0000 Min. : 0.000 Min. : 0.000
## 1st Qu.: 0.000 1st Qu.: 0.0000 1st Qu.: 0.000 1st Qu.: 0.000
## Median : 0.000 Median : 0.0000 Median : 1.000 Median : 0.000
## Mean : 1.022 Mean : 0.1484 Mean : 1.885 Mean : 2.094
## 3rd Qu.: 1.000 3rd Qu.: 0.0000 3rd Qu.: 3.000 3rd Qu.: 1.000
## Max. :95.000 Max. :25.0000 Max. :30.000 Max. :95.000
## NA's :1
## incarcerated man.divorced nsengimana murekatete
## Min. : 0.0000 Min. : 0.0000 Min. :0.0000 Min. : 0.0000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.:0.0000 1st Qu.: 0.0000
## Median : 0.0000 Median : 0.0000 Median :0.0000 Median : 0.0000
## Mean : 0.4347 Mean : 0.3367 Mean :0.3603 Mean : 0.3425
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000 3rd Qu.:0.0000 3rd Qu.: 1.0000
## Max. :95.0000 Max. :20.0000 Max. :8.0000 Max. :12.0000
##
## twahirwa mukandekezi nsabimana mukamana
## Min. : 0.0000 Min. :0.000 Min. : 0.0000 Min. : 0.0000
## 1st Qu.: 0.0000 1st Qu.:0.000 1st Qu.: 0.0000 1st Qu.: 0.0000
## Median : 0.0000 Median :0.000 Median : 0.0000 Median : 0.0000
## Mean : 0.2394 Mean :0.165 Mean : 0.4705 Mean : 0.4144
## 3rd Qu.: 0.0000 3rd Qu.:0.000 3rd Qu.: 1.0000 3rd Qu.: 1.0000
## Max. :10.0000 Max. :7.000 Max. :20.0000 Max. :15.0000
##
## ndayambaje nyiraneza bizimana nyirahabimana
## Min. : 0.0000 Min. : 0.0000 Min. : 0.0000 Min. : 0.000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.000
## Median : 0.0000 Median : 0.0000 Median : 0.0000 Median : 0.000
## Mean : 0.3296 Mean : 0.2685 Mean : 0.4331 Mean : 0.261
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000 3rd Qu.: 1.0000 3rd Qu.: 0.000
## Max. :30.0000 Max. :10.0000 Max. :12.0000 Max. :17.000
##
## ndagijimana mukandayisenga
## Min. : 0.0000 Min. : 0.0000
## 1st Qu.: 0.0000 1st Qu.: 0.0000
## Median : 0.0000 Median : 0.0000
## Mean : 0.3279 Mean : 0.2577
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000
## Max. :10.0000 Max. :20.0000
## Several populations, including widower,
male.community.health, teacher,
woman.smoke, muslim, and
incarcerated have maximum values that are very high. (It
turns out that 95 is the highest value that could be recorded during the
interviews; if respondents said that they were connected to more than 95
people in the group, the interviewers wrote 95 down.)
Now we use the topcode.data function to topcode all of
the responses at 30:
example.survey <- topcode.data(example.survey,
vars=known.popn.vars,
max=30)
## after topcoding: max. response for all popns is 30
summary(example.survey[,known.popn.vars])## widower nurse.or.doctor male.community.health teacher
## Min. : 0.0000 Min. : 0.0000 Min. : 0.000 Min. : 0.000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.000 1st Qu.: 0.000
## Median : 0.0000 Median : 0.0000 Median : 0.000 Median : 0.000
## Mean : 0.5831 Mean : 0.5062 Mean : 0.653 Mean : 1.216
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000 3rd Qu.: 1.000 3rd Qu.: 1.000
## Max. :30.0000 Max. :30.0000 Max. :30.000 Max. :30.000
##
## woman.smoke priest woman.gave.birth muslim
## Min. : 0.0000 Min. : 0.0000 Min. : 0.000 Min. : 0.000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.000 1st Qu.: 0.000
## Median : 0.0000 Median : 0.0000 Median : 1.000 Median : 0.000
## Mean : 0.9638 Mean : 0.1484 Mean : 1.885 Mean : 1.468
## 3rd Qu.: 1.0000 3rd Qu.: 0.0000 3rd Qu.: 3.000 3rd Qu.: 1.000
## Max. :30.0000 Max. :25.0000 Max. :30.000 Max. :30.000
## NA's :1
## incarcerated man.divorced nsengimana murekatete
## Min. : 0.0000 Min. : 0.0000 Min. :0.0000 Min. : 0.0000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.:0.0000 1st Qu.: 0.0000
## Median : 0.0000 Median : 0.0000 Median :0.0000 Median : 0.0000
## Mean : 0.3807 Mean : 0.3367 Mean :0.3603 Mean : 0.3425
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000 3rd Qu.:0.0000 3rd Qu.: 1.0000
## Max. :30.0000 Max. :20.0000 Max. :8.0000 Max. :12.0000
##
## twahirwa mukandekezi nsabimana mukamana
## Min. : 0.0000 Min. :0.000 Min. : 0.0000 Min. : 0.0000
## 1st Qu.: 0.0000 1st Qu.:0.000 1st Qu.: 0.0000 1st Qu.: 0.0000
## Median : 0.0000 Median :0.000 Median : 0.0000 Median : 0.0000
## Mean : 0.2394 Mean :0.165 Mean : 0.4705 Mean : 0.4144
## 3rd Qu.: 0.0000 3rd Qu.:0.000 3rd Qu.: 1.0000 3rd Qu.: 1.0000
## Max. :10.0000 Max. :7.000 Max. :20.0000 Max. :15.0000
##
## ndayambaje nyiraneza bizimana nyirahabimana
## Min. : 0.0000 Min. : 0.0000 Min. : 0.0000 Min. : 0.000
## 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.0000 1st Qu.: 0.000
## Median : 0.0000 Median : 0.0000 Median : 0.0000 Median : 0.000
## Mean : 0.3296 Mean : 0.2685 Mean : 0.4331 Mean : 0.261
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000 3rd Qu.: 1.0000 3rd Qu.: 0.000
## Max. :30.0000 Max. :10.0000 Max. :12.0000 Max. :17.000
##
## ndagijimana mukandayisenga
## Min. : 0.0000 Min. : 0.0000
## 1st Qu.: 0.0000 1st Qu.: 0.0000
## Median : 0.0000 Median : 0.0000
## Mean : 0.3279 Mean : 0.2577
## 3rd Qu.: 0.0000 3rd Qu.: 0.0000
## Max. :10.0000 Max. :20.0000
## If you look at the help page for topcode.data, you’ll
see that it can also handle situations where the variables can take on
special codes for missing values, refusals, and so forth.
Estimating network sizes
Now that we have finished preparing the data, we turn to esimating
the sizes of each respondent’s personal network. To do this using the
known population estimator, we use the kp.degree.estimator
function:
d.hat <- kp.individual.estimator_(resp.data=example.survey,
known.populations=known.popn.vars,
total.kp.size=sum(kp.vec),
alter.popn.size=tot.pop.size)$dbar.Fcell.F## NOTE: Ignoring any rows with missingness on any of the report variables.
summary(d.hat)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.00 25.28 58.99 101.21 126.42 1904.69We can examine the results with a histogram
library(ggplot2) # we'll use qplot from ggplot2 for plots
theme_set(theme_minimal())
qplot(d.hat, binwidth=25)## Warning: `qplot()` was deprecated in ggplot2 3.4.0.
## This warning is displayed once per session.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.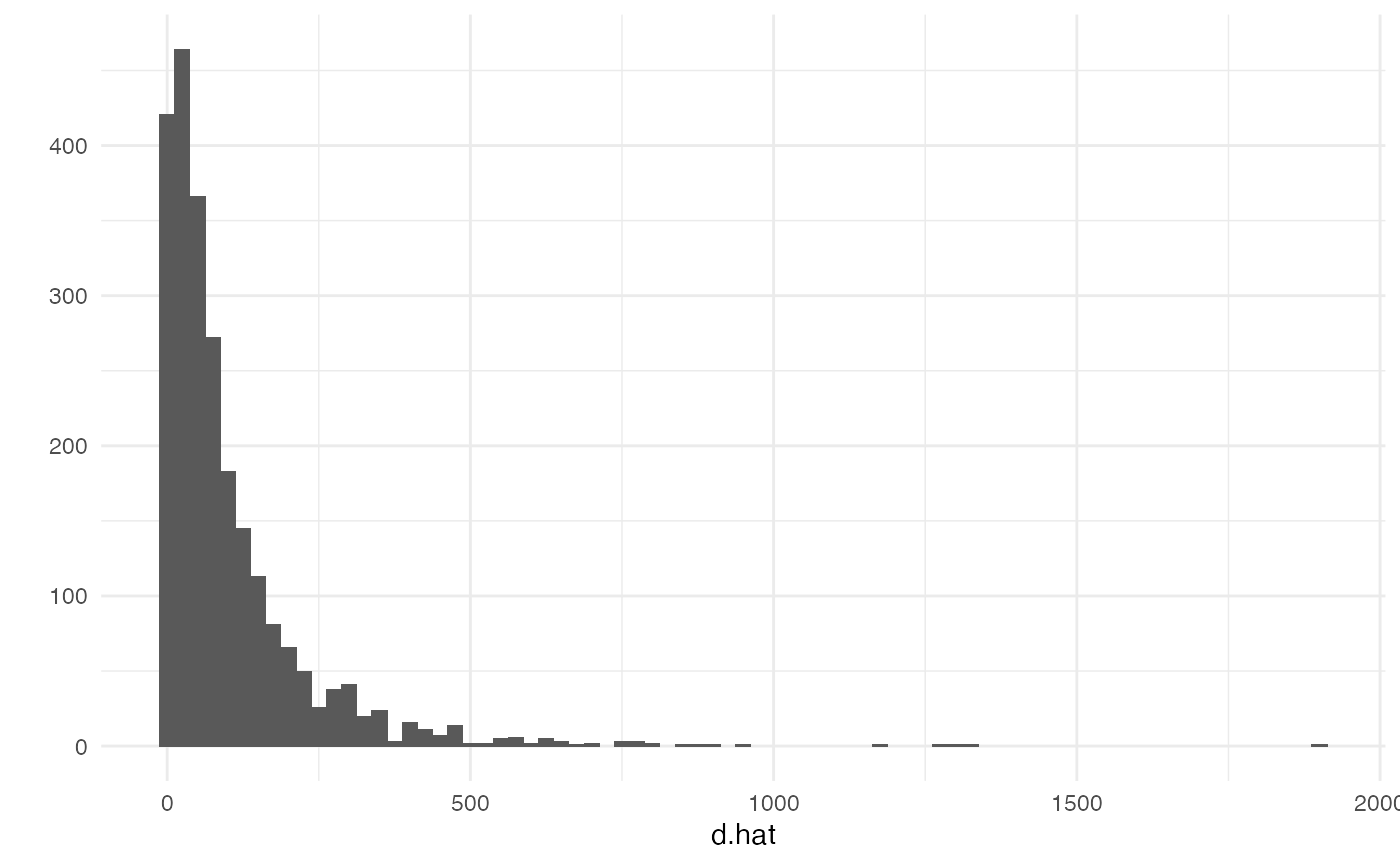
Now let’s append the degree estimates to the survey reports dataframe:
example.survey$d.hat <- d.hatEstimating hidden population size
Now that you have estimated degrees, you can use them to produce
estimates of the size of the hidden population. Here, we’ll take the
example of clients of female sex workers, clients
idu.est <- nsum.estimator(survey.data=example.survey,
d.hat.vals=d.hat,
total.popn.size=tot.pop.size,
y.vals="clients",
missing="complete.obs")Note that we had to specify that we should use only rows in our
dataset with no missing values through the
missing = "complete.obs" option, and also that we had to
pass in the total population size using the total.popn.size
option. The resulting estimate is
idu.est## $estimate
## [1] 47798.44
##
## $tot.connections
## [1] 1164
##
## $sum.d.hat
## [1] 243522.6This returns the estimate, and also the numerator and denominator used to compute it.
Variance estimation
In order to estimate the sampling uncertainty of our estimated totals, we can use the rescaled bootstrap technique; see Feehan and Salganik 2016 for more about the rescaled boostrap and how it can be applied to the network scale-up method. In order to use the rescaled boostrap, you need to be able to specify the sampling design of your study. In particular, you need to be able to describe the stratifcation (if any) and the primary sampling units used in the study.
idu.est <- bootstrap.estimates(## this describes the sampling design of the
## survey; here, the PSUs are given by the
## variable cluster, and the strata are given
## by the variable region
survey.design = ~ cluster + strata(region),
## the number of bootstrap resamples to obtain
## (NOTE: in practice, you should use more than 100.
## this keeps building the package relatively fast)
num.reps=100,
## this is the name of the function
## we want to use to produce an estimate
## from each bootstrapped dataset
estimator.fn="nsum.estimator",
## these are the sampling weights
weights="indweight",
## this is the name of the type of bootstrap
## we wish to use
bootstrap.fn="rescaled.bootstrap.sample",
## our dataset
survey.data=example.survey,
## other parameters we need to pass
## to the nsum.estimator function
d.hat.vals=d.hat,
total.popn.size=tot.pop.size,
y.vals="clients",
missing="complete.obs")## Warning: Unquoting language objects with `!!!` is deprecated as of rlang 0.4.0. Please
## use `!!` instead.
##
## # Bad: dplyr::select(data, !!!enquo(x))
##
## # Good: dplyr::select(data, !!enquo(x)) # Unquote single quosure
## dplyr::select(data, !!!enquos(x)) # Splice list of quosures
## This warning is displayed once every 8 hours.By default, bootstrap.estimates produces a list with
num.reps entries; each entry is the result of calling the
estimator function on one bootstrap resample.
Next, you can write a bit of code that will help us put all of these results together, for plotting and summarizing
library(plyr)
## combine the estimates together in one data frame
## (bootstrap.estimates gives us a list)
all.idu.estimates <- ldply(idu.est,
function(x) { data.frame(estimate=x$estimate) })We can examine the summarized results with a histogram or with
summarize.
## look at a histogram of the results
qplot(all.idu.estimates$estimate, binwidth=500)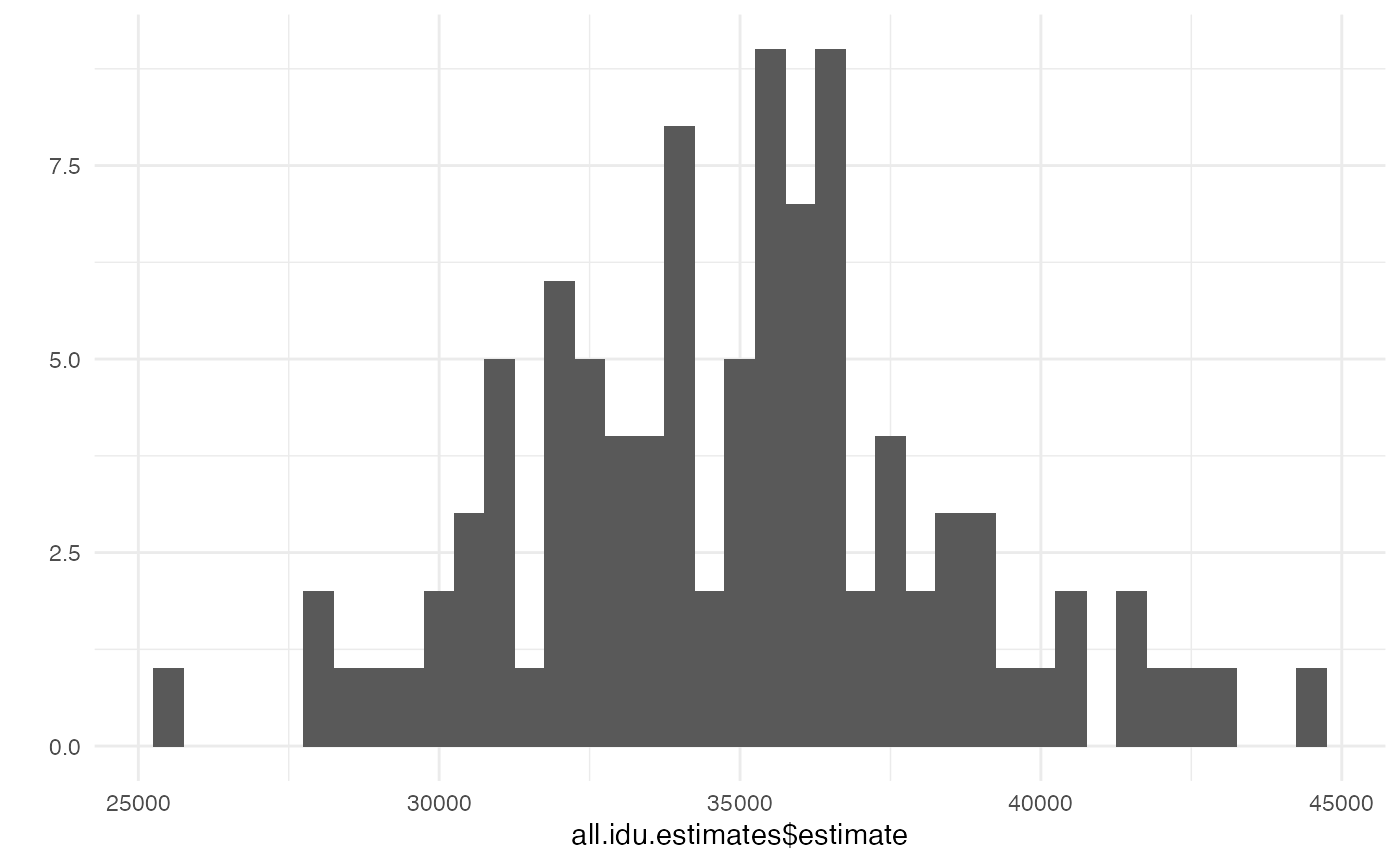
## summarize the results
summary(all.idu.estimates$estimate)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 27067 32759 35606 35465 37922 46234To produce 95% intervals using the percentile method you can do something like this
## 2.5% 97.5%
## 28318.34 43474.41Internal consistency checks
If you want to run internal consistency checks (see e.g. Feehan et al., 2016, Fig
3), you can use the nsum.internal.consistency function.
We specify that we wish to use only complete observations (ie, we will
remove rows that have any missing values from our calculations).
ic.result <- nsum.internal.consistency(survey.data=example.survey,
known.popns=kp.vec,
missing="complete.obs",
killworth.se=TRUE,
total.popn.size=tot.pop.size,
alter.popn.size=tot.pop.size,
kp.method=TRUE,
return.plot=TRUE)## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.
##
## NOTE: Ignoring any rows with missingness on any of the report variables.Now ic.result is a list that has a summary of the
results in the entry results
ic.result$results## name nsum.holdout.est known.size d.hat.sum killworth.se
## 1 widower 58708.19 36147 238978.6 1562.7564
## 2 nurse.or.doctor 51873.41 7807 234802.4 1482.4902
## 3 male.community.health 66955.68 22000 234632.8 1683.6063
## 4 teacher 128262.77 47745 228047.5 2356.3211
## 5 woman.smoke 93114.71 119438 249047.7 1924.5823
## 6 priest 14830.66 1004 240717.5 784.3396
## 7 woman.gave.birth 173204.79 256164 261828.8 2549.6303
## 8 muslim 137934.15 195449 255929.3 2305.5174
## 9 incarcerated 36619.80 68000 250137.9 1207.7348
## 10 man.divorced 32758.93 50698 247260.8 1149.1445
## 11 nsengimana 35697.55 32528 242873.8 1210.1866
## 12 murekatete 33933.74 30531 242826.2 1180.1303
## 13 twahirwa 23921.99 10420 240782.6 995.5567
## 14 mukandekezi 16382.94 10520 242325.2 821.5626
## 15 nsabimana 46399.77 48560 243966.7 1375.8868
## 16 mukamana 40565.23 51449 245777.0 1282.1053
## 17 ndayambaje 32841.38 22724 241463.7 1164.3150
## 18 nyiraneza 26637.59 21705 242514.4 1046.6453
## 19 bizimana 42949.20 38497 242612.2 1327.6590
## 20 nyirahabimana 25411.84 42727 247128.9 1012.7535
## 21 ndagijimana 32259.79 37375 244576.9 1146.6248
## 22 mukandayisenga 25249.25 35055 245551.8 1012.7533
## killworth.se.wgtdenom err abserr sqerr relerr
## 1 1562.7564 22561.191 22561.191 509007341 0.62415113
## 2 1482.4902 44066.413 44066.413 1941848743 5.64447456
## 3 1683.6063 44955.682 44955.682 2021013338 2.04344009
## 4 2356.3211 80517.771 80517.771 6483111367 1.68641262
## 5 1924.5823 -26323.291 26323.291 692915672 0.22039294
## 6 784.3396 13826.662 13826.662 191176573 13.77157538
## 7 2549.6303 -82959.207 82959.207 6882229975 0.32385193
## 8 2305.5174 -57514.848 57514.848 3307957792 0.29427036
## 9 1207.7348 -31380.200 31380.200 984716944 0.46147353
## 10 1149.1445 -17939.074 17939.074 321810359 0.35384184
## 11 1210.1866 3169.553 3169.553 10046064 0.09744075
## 12 1180.1303 3402.735 3402.735 11578609 0.11145182
## 13 995.5567 13501.990 13501.990 182303747 1.29577644
## 14 821.5626 5862.944 5862.944 34374109 0.55731404
## 15 1375.8868 -2160.232 2160.232 4666603 0.04448583
## 16 1282.1053 -10883.772 10883.772 118456503 0.21154488
## 17 1164.3150 10117.380 10117.380 102361383 0.44522884
## 18 1046.6453 4932.592 4932.592 24330468 0.22725604
## 19 1327.6590 4452.195 4452.195 19822041 0.11565044
## 20 1012.7535 -17315.161 17315.161 299814807 0.40525104
## 21 1146.6248 -5115.214 5115.214 26165412 0.13686191
## 22 1012.7533 -9805.749 9805.749 96152722 0.27972470Since we passed the argument return.plot=TRUE to the
function, we also get a plot:
print(ic.result$plot)
This plot is a ggplot2 object, so we can customize it if
we want. As a very simple example, we can change the title:
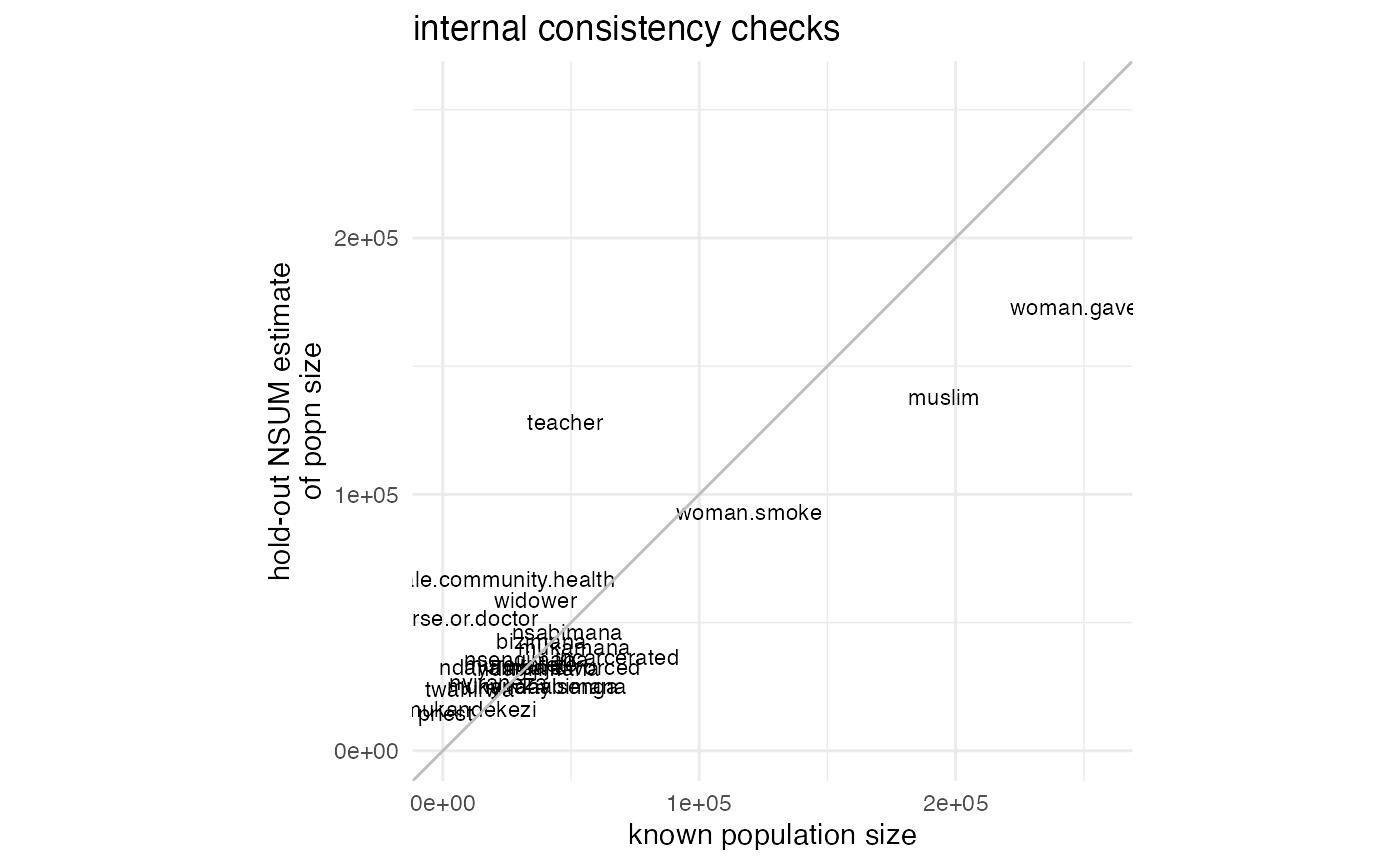
The ggplot2 website has more information on modifying ggplot2 objects.